| GNMT | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | GNMT, HEL-S-182mP, glycine N-methyltransferase | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 606628; MGI: 1202304; HomoloGene: 7741; GeneCards: GNMT; OMA:GNMT - orthologs | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Glycine N-methyltransferase is an enzyme that in humans is encoded by the GNMT gene.
Discovery
The enzyme was first described by Blumenstein and Williams (1960) in guinea pig liver. However, this enzyme was not purified until 1972 in the rabbit liver by Kerr. In 1984, Cook and Wagner demonstrated that a liver cytosolic folate binding protein is identical to GNMT. The human GMNT gene was cloned in 2000 by Chen and coworkers.
Tissue distribution
GNMT is an abundant enzyme in liver cytosol and consists of 0.9% to 3% of the soluble protein present in liver. In addition to liver, GNMT activity has been found in a number of other tissues including pancreas and kidney. GNMT is most abundant in the peri-portal region of the liver and exocrine tissue of the pancreas. The GNMT proteins located in tissues that are actively in secretion, such as the proximal kidney tubules, the submaxillary glands and the intestinal mucosa. GNMT is also expressed in various neurons presented in the cerebral cortex, hippocampus, substantia nigra and cerebellum. The presence of GNMT in these cells suggests that this enzyme may play a role in secretion.
Structure
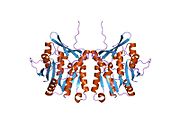
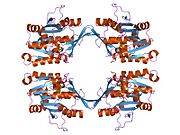
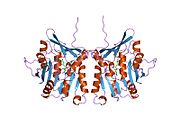
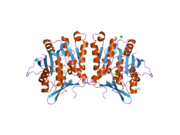
The properties of GNMT protein from rabbits, rats and humans, either purified from liver/pancreas, or expressed in Escherichia coli, have been well characterized. All GNMTs have very similar molecular and kinetic properties. Comparison of the cDNA and protein sequences of human, rabbit, pig and rat GNMTs shows similarities of over 84% at the nucleotide level and about 90% at the amino acid level. All GNMTs are 130 kDa tetramers consisting of four identical subunits, each having a Mr of 32 kDa. The structure of recombinant rat, mouse and human GNMTs have been solved. The four nearly spherical subunits are arranged to form a flat and square tetramer with a large hole in the center. The active sites are located in the near center of each subunit.
Function
Glycine N-methyltransferase catalyzes the synthesis of N-methylglycine (sarcosine) from glycine using S-adenosylmethionine (SAM) (AdoMet) as the methyl donor. GNMT acts as an enzyme to regulate the ratio of S-adenosylmethionine (SAM) to S-adenosylhomocysteine (SAH) (AdoHcy) and participates in the detoxification pathway in liver cells. GNMT competes with tRNA methyltransferases for SAM and the product, S-adenosylhomocysteine (SAH), is a potent inhibitor of tRNA methyltransferases and a relatively weak inhibitor of GNMT. GNMT regulates the relative levels of SAM and SAH. Since SAM is the methyl donor for almost all cellular methylation reactions. GNMT is therefore likely to regulate cellular methylation capacity. An endogenous ligand of GNMT, 5-methyltetrahydropteroylpentaglutamate (5-CH3-H4PteGIu5) is a powerful inhibitor of this enzyme. Thus, GNMT has been proposed to link the de novo synthesis of methyl groups to the ratio of SAM to SAH, which in turn serves as a bridge between methionine and one-carbon metabolism.
In addition to the methyltransferase activity, the 4S polycyclic aromatic hydrocarbon (PAH)-binding protein and GNMT are one and the same protein. The catalytic site resembles a molecular basket, unlike most other SAM-dependent methyltransferases, which therefore suggests that GNMT may be capable of capturing unidentified chemicals as a part of a detoxification process. Therefore, GNMT has been proposed to be a protein with diverse functionality.
Clinical significance
GNMT has been shown to detoxify some environmental carcinogens such as polyaromatic hydrocarbons and aflatoxin.
There is mounting evidence that supports the involvement of GNMT deficiency in liver carcinogenesis.
Inducer
The glycoside natural product 1,2,3,4,6-penta-O-galloyl-β-d-glucopyranoside (PGG) isolated from Paeonia lactiflora, an Asian flower plant, induces GNMT mRNA and protein expression in Huh7 human hepatoma cells.
References
- ^ GRCh38: Ensembl release 89: ENSG00000124713 – Ensembl, May 2017
- ^ GRCm38: Ensembl release 89: ENSMUSG00000002769 – Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Chen YM, Shiu JY, Tzeng SJ, Shih LS, Chen YJ, Lui WY, Chen PH (March 1998). "Characterization of glycine-N-methyltransferase-gene expression in human hepatocellular carcinoma". International Journal of Cancer. 75 (5): 787–93. doi:10.1002/(SICI)1097-0215(19980302)75:5<787::AID-IJC20>3.0.CO;2-2. PMID 9495250. S2CID 42285102.
- ^ Chen YM, Chen LY, Wong FH, Lee CM, Chang TJ, Yang-Feng TL (May 2000). "Genomic structure, expression, and chromosomal localization of the human glycine N-methyltransferase gene". Genomics. 66 (1): 43–7. doi:10.1006/geno.2000.6188. PMID 10843803.
- ^ "Entrez Gene: GNMT glycine N-methyltransferase".
- Blumenstein J, Williams GR (September 1960). "The enzymic N-methylation of glycine". Biochemical and Biophysical Research Communications. 3 (3): 259–263. doi:10.1016/0006-291X(60)90235-7.
- ^ Kerr SJ, Borek E (1972). "The tRNA methyltransferases". Advances in Enzymology and Related Areas of Molecular Biology. Vol. 36. pp. 1–27. doi:10.1002/9780470122815.ch1. ISBN 9780470122815. PMID 4563428.
- Cook RJ, Wagner C (1984). "Glycine N-methyltransferase is a folate binding protein of rat liver cytosol". Proceedings of the National Academy of Sciences of the United States of America. 81 (12): 3631–4. Bibcode:1984PNAS...81.3631C. doi:10.1073/pnas.81.12.3631. PMC 345272. PMID 6587377.
- ^ Yeo EJ, Wagner C (1994). "Tissue distribution of glycine N-methyltransferase, a major folate-binding protein of liver". Proceedings of the National Academy of Sciences of the United States of America. 91 (1): 210–4. Bibcode:1994PNAS...91..210Y. doi:10.1073/pnas.91.1.210. PMC 42916. PMID 8278367.
- Yang CP, Wang HA, Tsai TH, Fan A, Hsu CL, Chen CJ, Hong CJ, Chen YM (August 2012). "Characterization of the neuropsychological phenotype of glycine N-methyltransferase-/- mice and evaluation of its responses to clozapine and sarcosine treatments". European Neuropsychopharmacology. 22 (8): 596–606. doi:10.1016/j.euroneuro.2011.12.007. PMID 22264868. S2CID 11604802.
- Heady JE, Kerr SJ (1973). "Purification and characterization of glycine N-methyltransferase". The Journal of Biological Chemistry. 248 (1): 69–72. doi:10.1016/S0021-9258(19)44446-3. PMID 4692843.
- Ogawa H, Fujioka M (1982). "Purification and properties of glycine N-methyltransferase from rat liver". The Journal of Biological Chemistry. 257 (7): 3447–52. doi:10.1016/S0021-9258(18)34798-7. PMID 6801046.
- ^ Ogawa H, Gomi T, Fujioka M (1993). "Mammalian glycine N-methyltransferases. Comparative kinetic and structural properties of the enzymes from human, rat, rabbit and pig livers". Comparative Biochemistry and Physiology. B, Comparative Biochemistry. 106 (3): 601–11. doi:10.1016/0305-0491(93)90137-t. PMID 8281755.
- Yeo EJ, Wagner C (1992). "Purification and properties of pancreatic glycine N-methyltransferase". The Journal of Biological Chemistry. 267 (34): 24669–74. doi:10.1016/S0021-9258(18)35816-2. PMID 1332963.
- ^ Fu Z, Hu Y, Konishi K, Takata Y, Ogawa H, Gomi T, Fujioka M, Takusagawa F (1996). "Crystal structure of glycine N-methyltransferase from rat liver". Biochemistry. 35 (37): 11985–93. doi:10.1021/bi961068n. PMID 8810903.
- Pakhomova S, Luka Z, Grohmann S, Wagner C, Newcomer ME (2004). "Glycine N-methyltransferases: a comparison of the crystal structures and kinetic properties of recombinant human, mouse and rat enzymes". Proteins. 57 (2): 331–7. doi:10.1002/prot.20209. PMID 15340920. S2CID 26065465.
- ^ Luka Z, Mudd SH, Wagner C (2009). "Glycine N-methyltransferase and regulation of S-adenosylmethionine levels". The Journal of Biological Chemistry. 284 (34): 22507–11. doi:10.1074/jbc.R109.019273. PMC 2755656. PMID 19483083.
- McCabe DC, Caudill MA (2005). "DNA methylation, genomic silencing, and links to nutrition and cancer". Nutrition Reviews. 63 (6 Pt 1): 183–95. doi:10.1111/j.1753-4887.2005.tb00136.x. PMID 16028562.
- ^ Wagner C, Briggs WT, Cook RJ (1985). "Inhibition of glycine N-methyltransferase activity by folate derivatives: implications for regulation of methyl group metabolism". Biochemical and Biophysical Research Communications. 127 (3): 746–52. doi:10.1016/s0006-291x(85)80006-1. PMID 3838667.
- Raha A, Wagner C, MacDonald RG, Bresnick E (1994). "Rat liver cytosolic 4 S polycyclic aromatic hydrocarbon-binding protein is glycine N-methyltransferase". The Journal of Biological Chemistry. 269 (8): 5750–6. doi:10.1016/S0021-9258(17)37525-7. PMID 8119914.
- Bhat R, Bresnick E (August 1997). "Glycine N-methyltransferase is an example of functional diversity. Role as a polycyclic aromatic hydrocarbon-binding receptor". The Journal of Biological Chemistry. 272 (34): 21221–6. doi:10.1074/jbc.272.34.21221. PMID 9261130.
- Yen CH, Lin YT, Chen HL, Chen SY, Chen YM (January 2013). "The multi-functional roles of GNMT in toxicology and cancer". Toxicology and Applied Pharmacology. 266 (1): 67–75. doi:10.1016/j.taap.2012.11.003. PMID 23147572.
- Barić I (2009). "Inherited disorders in the conversion of methionine to homocysteine". Journal of Inherited Metabolic Disease. 32 (4): 459–71. doi:10.1007/s10545-009-1146-4. PMID 19585268. S2CID 8659319.
- Kant R, Yen CH, Lu CK, Lin YC, Li JH, Chen YM (May 2016). "Identification of 1,2,3,4,6-Penta-O-galloyl-β-d-glucopyranoside as a Glycine N-Methyltransferase Enhancer by High-Throughput Screening of Natural Products Inhibits Hepatocellular Carcinoma". International Journal of Molecular Sciences. 17 (5): 669. doi:10.3390/ijms17050669. PMC 4881495. PMID 27153064.
Further reading
- Chen M, Huang YL, Huang YC, Shui IM, Giovannucci E, Chen YC, Chen YM (May 2014). "Genetic polymorphisms of the glycine N-methyltransferase and prostate cancer risk in the health professionals follow-up study". PLOS ONE. 9 (5): e94683. Bibcode:2014PLoSO...994683C. doi:10.1371/journal.pone.0094683. PMC 4011739. PMID 24800880.
- Chou WY, Zhao JF, Chen YM, Lee KI, Su KH, Shyue SK, Lee TS (March 2014). "Role of glycine N-methyltransferase in experimental ulcerative colitis". Journal of Gastroenterology and Hepatology. 29 (3): 494–501. doi:10.1111/jgh.12434. PMID 24219143. S2CID 22074228.
- Wang YC, Lin WL, Lin YJ, Tang FY, Chen YM, Chiang EP (February 2014). "A novel role of the tumor suppressor GNMT in cellular defense against DNA damage". International Journal of Cancer. 134 (4): 799–810. doi:10.1002/ijc.28420. PMID 23922098. S2CID 24507645.
- Chen YM, Liao YJ (July 2013). "Phosphorylated forms of GNMT in mouse liver". Journal of Proteomics. 87: 132–3. doi:10.1016/j.jprot.2013.01.007. PMID 23340451.
- Chen CY, Ching LC, Liao YJ, Yu YB, Tsou CY, Shyue SK, Chen YM, Lee TS (July 2012). "Deficiency of glycine N-methyltransferase aggravates atherosclerosis in apolipoprotein E-null mice". Molecular Medicine. 18 (5): 744–52. doi:10.2119/molmed.2011.00396. PMC 3409278. PMID 22415010.
- Liao YJ, Chen TL, Lee TS, Wang HA, Wang CK, Liao LY, Liu RS, Huang SF, Chen YM (May 2012). "Glycine N-methyltransferase deficiency affects Niemann-Pick type C2 protein stability and regulates hepatic cholesterol homeostasis". Molecular Medicine. 18 (3): 412–22. doi:10.2119/molmed.2011.00258. PMC 3356423. PMID 22183894.
- Chen M, Ho CW, Huang YC, Wu KY, Wu MT, Jeng HA, Chen CJ, Shih TS, Lai CH, Pan CH, Chen YM (July 2011). "Glycine N-methyltransferase affects urinary 1-hydroxypyrene and 8-hydroxy-2'-deoxyguanosine levels after PAH exposure". Journal of Occupational and Environmental Medicine. 53 (7): 812–9. doi:10.1097/JOM.0b013e318222b79a. PMID 21691217. S2CID 205722774.
- Wang YC, Tang FY, Chen SY, Chen YM, Chiang EP (May 2011). "Glycine-N methyltransferase expression in HepG2 cells is involved in methyl group homeostasis by regulating transmethylation kinetics and DNA methylation". The Journal of Nutrition. 141 (5): 777–82. doi:10.3945/jn.110.135954. PMID 21411609.
- Wang YC, Chen YM, Lin YJ, Liu SP, Chiang EP (May–Jun 2011). "GNMT expression increases hepatic folate contents and folate-dependent methionine synthase-mediated homocysteine remethylation". Molecular Medicine. 17 (5–6): 486–94. doi:10.2119/molmed.2010.00243. PMC 3105148. PMID 21210071.
- Liao YJ, Chen KH, Huang SF, Chen TL, Wang CK, Chien CH, Tsai TF, Liu SP, Chen YM (April 2010). "Deficiency of glycine N-methyltransferase results in deterioration of cellular defense to stress in mouse liver". Proteomics: Clinical Applications. 4 (4): 394–406. doi:10.1002/prca.200900074. PMID 21137059. S2CID 8638838.
- Tsai MJ, Chen YM, Weng CF, Liou DY, Yang HC, Chen CH, Liao RI, Kuo FS, Chiu CW, Kuo HS, Huang MC, Lin YL, Lee MJ, Kuo WC, Huang WC, Cheng H (June 2010). "Enhanced expression of glycine N-methyltransferase by adenovirus-mediated gene transfer in CNS culture is neuroprotective". Annals of the New York Academy of Sciences. 1199 (1): 194–203. Bibcode:2010NYASA1199..194T. doi:10.1111/j.1749-6632.2009.05169.x. PMID 20633125. S2CID 20120622.
- Chen YM, Liao YJ, Liu SP, Tsai TF (June 2009). "Phenotypic differences between two Gnmt-/- mouse models for hepatocellular carcinoma". Hepatology. 49 (6): 2130–1, author reply 2131. doi:10.1002/hep.22984. PMID 19475685. S2CID 205869970.
- Lee CM, Shih YP, Wu CH, Chen YM (August 2009). "Characterization of the 5' regulatory region of the human Glycine N-methyltransferase gene". Gene. 443 (1–2): 151–7. doi:10.1016/j.gene.2009.05.001. PMID 19439180.
- Huang YC, Chen M, Shyr YM, Su CH, Chen CK, Li AF, Ho DM, Chen YM (September 2008). "Glycine N-methyltransferase is a favorable prognostic marker for human cholangiocarcinoma". Journal of Gastroenterology and Hepatology. 23 (9): 1384–9. doi:10.1111/j.1440-1746.2008.05488.x. PMID 18624901. S2CID 24227544.
- Huang YC, Lee CM, Chen M, Chung MY, Chang YH, Huang WJ, Ho DM, Pan CC, Wu TT, Yang S, Lin MW, Hsieh JT, Chen YM (March 2007). "Haplotypes, loss of heterozygosity, and expression levels of glycine N-methyltransferase in prostate cancer". Clinical Cancer Research. 13 (5): 1412–20. doi:10.1158/1078-0432.CCR-06-1551. PMID 17332283.
- Lee CM, Chen SY, Lee YC, Huang CY, Chen YM (July 2006). "Benzopyrene and glycine N-methyltransferse interactions: gene expression profiles of the liver detoxification pathway". Toxicology and Applied Pharmacology. 214 (2): 126–35. doi:10.1016/j.taap.2005.12.020. PMID 16545412.
- Chen SY, Lin JR, Darbha R, Lin P, Liu TY, Chen YM (May 2004). "Glycine N-methyltransferase tumor susceptibility gene in the benzo(a)pyrene-detoxification pathway". Cancer Research. 64 (10): 3617–23. doi:10.1158/0008-5472.CAN-03-3726. PMID 15150120.
- Liu HH, Chen KH, Shih YP, Lui WY, Wong FH, Chen YM (Jan–Feb 2003). "Characterization of reduced expression of glycine N-methyltransferase in cancerous hepatic tissues using two newly developed monoclonal antibodies". Journal of Biomedical Science. 10 (1): 87–97. doi:10.1007/BF02256001. PMID 12566990.
- Tseng TL, Shih YP, Huang YC, Wang CK, Chen PH, Chang JG, Yeh KT, Chen YM, Buetow KH (February 2003). "Genotypic and phenotypic characterization of a putative tumor susceptibility gene, GNMT, in liver cancer". Cancer Research. 63 (3): 647–54. PMID 12566309.
- Lee CM, Yen CH, Tzeng TY, Huang YZ, Chou KH, Chang TJ, Arthur Chen YM (September 2013). "Androgen response element of the glycine N-methyltransferase gene is located in the coding region of its first exon". Bioscience Reports. 33 (5): 761–769. doi:10.1042/BSR20130030. PMC 3775523. PMID 23883094.
- Li CH, Lin MH, Chu SH, Tu PH, Fang CC, Yen CH, Liang PI, Huang JC, Su YC, Sytwu HK, Chen YM (March 2015). "Role of glycine N-methyltransferase in the regulation of T-cell responses in experimental autoimmune encephalomyelitis". Molecular Medicine. 20 (1): 684–96. doi:10.2119/molmed.2014.00133. PMC 4398670. PMID 25535034.
- Yen CH, Hung JH, Ueng YF, Liu SP, Chen SY, Liu HH, Chou TY, Tsai TF, Darbha R, Hsieh LL, Chen YM (March 2009). "Glycine N-methyltransferase affects the metabolism of aflatoxin B1 and blocks its carcinogenic effect". Toxicology and Applied Pharmacology. 235 (3): 296–304. doi:10.1016/j.taap.2008.12.013. PMID 19146867.
- Chen YM, Shiu JY, Tzeng SJ, Shih LS, Chen YJ, Lui WY, Chen PH (March 1998). "Characterization of glycine-N-methyltransferase-gene expression in human hepatocellular carcinoma". International Journal of Cancer. 75 (5): 787–93. doi:10.1002/(sici)1097-0215(19980302)75:5<787::aid-ijc20>3.0.co;2-2. PMID 9495250. S2CID 42285102.
- Wagner C, Decha-Umphai W, Corbin J (June 1989). "Phosphorylation modulates the activity of glycine N-methyltransferase, a folate binding protein. In vitro phosphorylation is inhibited by the natural folate ligand". The Journal of Biological Chemistry. 264 (16): 9638–42. doi:10.1016/S0021-9258(18)60578-2. PMID 2722853.
- Wagner C, Decha-Umphai W, Corbin J (June 1989). "Phosphorylation modulates the activity of glycine N-methyltransferase, a folate binding protein. In vitro phosphorylation is inhibited by the natural folate ligand". The Journal of Biological Chemistry. 264 (16): 9638–42. doi:10.1016/S0021-9258(18)60578-2. PMID 2722853.
- Mudd SH, Ebert MH, Scriver CR (August 1980). "Labile methyl group balances in the human: the role of sarcosine". Metabolism. 29 (8): 707–20. doi:10.1016/0026-0495(80)90192-4. PMID 6157075.
- Mudd SH, Cerone R, Schiaffino MC, Fantasia AR, Minniti G, Caruso U, Lorini R, Watkins D, Matiaszuk N, Rosenblatt DS, Schwahn B, Rozen R, LeGros L, Kotb M, Capdevila A, Luka Z, Finkelstein JD, Tangerman A, Stabler SP, Allen RH, Wagner C (August 2001). "Glycine N-methyltransferase deficiency: a novel inborn error causing persistent isolated hypermethioninaemia". Journal of Inherited Metabolic Disease. 24 (4): 448–64. doi:10.1023/A:1010577512912. PMID 11596649. S2CID 28155061.
- Luka Z, Cerone R, Phillips JA, Mudd HS, Wagner C (January 2002). "Mutations in human glycine N-methyltransferase give insights into its role in methionine metabolism". Human Genetics. 110 (1): 68–74. doi:10.1007/s00439-001-0648-4. PMID 11810299. S2CID 9819456.
- Tseng TL, Shih YP, Huang YC, Wang CK, Chen PH, Chang JG, Yeh KT, Chen YM, Buetow KH (February 2003). "Genotypic and phenotypic characterization of a putative tumor susceptibility gene, GNMT, in liver cancer". Cancer Research. 63 (3): 647–54. PMID 12566309.
- Møller MT, Samari HR, Fengsrud M, Strømhaug PE, øStvold AC, Seglen PO (July 2003). "Okadaic acid-induced, naringin-sensitive phosphorylation of glycine N-methyltransferase in isolated rat hepatocytes". The Biochemical Journal. 373 (Pt 2): 505–13. doi:10.1042/BJ20030502. PMC 1223502. PMID 12697024.
- Luka Z, Wagner C (December 2003). "Effect of naturally occurring mutations in human glycine N-methyltransferase on activity and conformation". Biochemical and Biophysical Research Communications. 312 (4): 1067–72. doi:10.1016/j.bbrc.2003.11.037. PMID 14651980.
- Augoustides-Savvopoulou P, Luka Z, Karyda S, Stabler SP, Allen RH, Patsiaoura K, Wagner C, Mudd SH (2004). "Glycine N -methyltransferase deficiency: a new patient with a novel mutation". Journal of Inherited Metabolic Disease. 26 (8): 745–59. doi:10.1023/B:BOLI.0000009978.17777.33. PMID 14739680. S2CID 9179123.
- Beagle B, Yang TL, Hung J, Cogger EA, Moriarty DJ, Caudill MA (December 2005). "The glycine N-methyltransferase (GNMT) 1289 C->T variant influences plasma total homocysteine concentrations in young women after restricting folate intake". The Journal of Nutrition. 135 (12): 2780–5. doi:10.1093/jn/135.12.2780. PMID 16317120.
- Luka Z, Pakhomova S, Luka Y, Newcomer ME, Wagner C (September 2007). "Destabilization of human glycine N-methyltransferase by H176N mutation". Protein Science. 16 (9): 1957–64. doi:10.1110/ps.072921507. PMC 2206963. PMID 17660255.