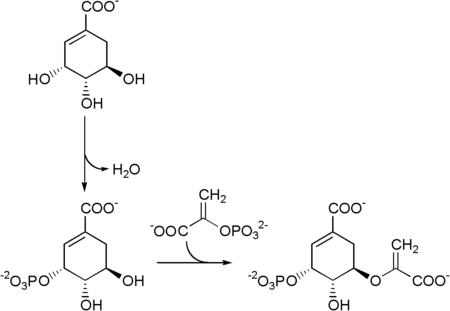
The shikimate pathway (shikimic acid pathway) is a seven-step metabolic pathway used by bacteria, archaea, fungi, algae, some protozoans, and plants for the biosynthesis of folates and aromatic amino acids (tryptophan, phenylalanine, and tyrosine). This pathway is not found in mammals.
The five enzymes involved in the shikimate pathway are 3-dehydroquinate dehydratase, shikimate dehydrogenase, shikimate kinase, EPSP synthase, and chorismate synthase. In bacteria and eurkaryotes, the pathway starts with two substrates, phosphoenol pyruvate and erythrose-4-phosphate, are processed by DAHP synthase and 3-dehydroquinate synthase to form 3-dehydroquinate. In archaea, 2-amino-3,7-dideoxy-D-threo-hept-6-ulosonate synthase condenses L-Aspartic-4-semialdehyde with a sugar to form 2-amino-3,7-dideoxy-D-threo-hept-6-ulosonate, which is then turned by 3-dehydroquinate synthase II into 3-dehydroquinate. Both pathways end with chorismate (chrorismic acid), a substrate for the three aromatic amino acids. The fifth enzyme involved is the shikimate kinase, an enzyme that catalyzes the ATP-dependent phosphorylation of shikimate to form shikimate 3-phosphate (shown in the figure below). Shikimate 3-phosphate is then coupled with phosphoenol pyruvate to give 5-enolpyruvylshikimate-3-phosphate via the enzyme 5-enolpyruvylshikimate-3-phosphate (EPSP) synthase. Glyphosate, the herbicidal ingredient in Roundup, is a competitive inhibitor of EPSP synthase, acting as a transition state analog that binds more tightly to the EPSPS-S3P complex than PEP and inhibits the shikimate pathway.
Then 5-enolpyruvylshikimate-3-phosphate is transformed into chorismate by a chorismate synthase.
Prephenic acid is then synthesized by a Claisen rearrangement of chorismate by chorismate mutase.
Prephenate is oxidatively decarboxylated with retention of the hydroxyl group to give p-hydroxyphenylpyruvate, which is transaminated using glutamate as the nitrogen source to give tyrosine and α-ketoglutarate.
References
- Morar, Mariya; White, Robert H.; Ealick, Steven E. (1 September 2007). "Structure of 2-Amino-3,7-dideoxy- d - threo -hept-6-ulosonic Acid Synthase, a Catalyst in the Archaeal Pathway for the Biosynthesis of Aromatic Amino Acids ,". Biochemistry. 46 (37): 10562–10571. doi:10.1021/bi700934v.
- Herrmann, K. M.; Weaver, L. M. (1999). "The Shikimate Pathway". Annual Review of Plant Physiology and Plant Molecular Biology. 50: 473–503. doi:10.1146/annurev.arplant.50.1.473. PMID 15012217.
- Helmut Goerisch (1978). "On the mechanism of the chorismate mutase reaction". Biochemistry. 17 (18): 3700–3705. doi:10.1021/bi00611a004. PMID 100134.
- Peter Kast; Yadu B. Tewari; Olaf Wiest; Donald Hilvert; Kendall N. Houk; Robert N. Goldberg (1997). "Thermodynamics of the Conversion of Chorismate to Prephenate: Experimental Results and Theoretical Predictions". J. Phys. Chem. B. 101 (50): 10976–10982. doi:10.1021/jp972501l.
Bibliography
- Edwin Haslam (1993). Shikimic Acid: Metabolism and Metabolites (1st ed.). ISBN 0471939994.
- Brown, Stewart A.; Neish, A. C. (1955). "Shikimic Acid as a Precursor in Lignin Biosynthesis". Nature. 175 (4459): 688–689. Bibcode:1955Natur.175..688B. doi:10.1038/175688a0. ISSN 0028-0836. PMID 14370198. S2CID 4273320.
- Weinstein, L. H.; Porter, C. A.; Laurencot, H. J. (1962). "Role of the Shikimic Acid Pathway in the Formation of Tryptophan in Higher Plants : Evidence for an Alternative Pathway in the Bean". Nature. 194 (4824): 205–206. Bibcode:1962Natur.194..205W. doi:10.1038/194205a0. ISSN 0028-0836. S2CID 4160308.
- Wilson, D J; Patton, S; Florova, G; Hale, V; Reynolds, K A (1998). "The shikimic acid pathway and polyketide biosynthesis". Journal of Industrial Microbiology and Biotechnology. 20 (5): 299–303. doi:10.1038/sj.jim.2900527. ISSN 1367-5435. S2CID 41117722.