 | |
| Names | |
|---|---|
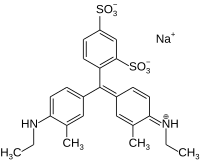
| Preferred IUPAC name Sodium 4-{(Z)-methyl}-3-sulfobenzene-1-sulfonate | |
| Other names
Acid Blue 147 xylene cyanole xylene cyanol FF xylene cyanole FF C.I. 42135 | |
| Identifiers | |
| CAS Number | |
| 3D model (JSmol) | |
| ChemSpider | |
| ECHA InfoCard | 100.018.334 |
| EC Number |
|
| PubChem CID | |
| CompTox Dashboard (EPA) | |
InChI
| |
SMILES
| |
| Properties | |
| Chemical formula | C25H27N2NaO6S2 |
| Molar mass | 538.61 g·mol |
| Hazards | |
| GHS labelling: | |
| Pictograms | 
|
| Signal word | Warning |
| Hazard statements | H315, H319, H335 |
| Precautionary statements | P261, P264, P271, P280, P302+P352, P304+P340, P305+P351+P338, P312, P321, P332+P313, P337+P313, P362, P403+P233, P405, P501 |
| Except where otherwise noted, data are given for materials in their standard state (at 25 °C , 100 kPa).
| |
Xylene cyanol can be used as an electrophoretic color marker, or tracking dye, to monitor the process of agarose gel electrophoresis and polyacrylamide gel electrophoresis. Bromophenol blue and orange G can also be used for this purpose.
Once mixed with the sample, the concentration of xylene cyanol is typically about 0.005% to 0.03%.
Migration speed
In 1% agarose gels, xylene cyanol migrates at about the same rate as a 4 to 5 kilobase pair DNA fragment, although this depends on the buffer used. Xylene cyanol on a 6% polyacrylamide gel migrates at the speed of a 140 base pair DNA fragment. On 20% denaturating (7 M urea) polyacrylamide gel electrophoresis (PAGE), xylene cyanol migrates at about the rate of 25 bases oligonucleotide.
References
- Lela Buckingham and Maribeth L. Flaws (2007). Molecular Diagnostics: Fundamentals, Methods, & Clinical Applications. F.A. Davis Company. p. 91. ISBN 9780803616592.